JupyterLab#
With this app you can write and run Jupyter notebooks containing annotated Python, R or Julia code (among other languages). IPython consoles are available as well. One of the benefits of JupyterLab is that it supports different types of user-defined environments, as will become clear below.
The top-level notebook directory is the selected ‘Working directory’ in
the resources form. Note however, if you have set c.ServerApp.root_dir
in your Jupyter configuration file
(default = ~/.jupyter/jupyter_server_config.py), this config will take
priority over your ‘Working directory’ in the resources form.
Pure module environment#
In the resources form, besides the resources that are shared between all apps, you can also choose between different Toolchain and Python versions from a drop-down menu. Based on that choice, the corresponding JupyterLab module will be loaded together with its dependencies.
Furthermore, you may choose to tick one of the checkboxes to load popular
modules from the same toolchain, such as SciPy-bundle (for widely used
packages like scipy, numpy, pandas and more) and/or matplotlib.
Once you launch a JupyterLab session, a default kernel called Python 3
(ipykernel) is already available in your session. This kernel, in addition to
the Python standard library, would enable using extra packages from
SciPy-bundle and/or matplotlib, if you selected them in the resources
form.
If the selected modules do not provide all Python packages that you need, you
can load extra modules with Python packages via module load commands in the
‘Pre-run Scriptlet’ of the resources form.
Warning
If you use JupyterLab in the pure module environment, remember to be consistent in your choice of toolchain version as this determines the versions of Python and Python packages that will be used.
Jupyter kernels#
The following table shows the kernels available in JupyterLab and the corresponding modules that have to be loaded to enable them:
Kernel |
Software Module |
|---|---|
Python |
(loaded by default) |
R |
|
Julia |
|
The default lab environment only loads the Python kernel upon launch. You can activate any other kernel by loading its corresponding software module. Once a module providing a new kernel is loaded, a new icon will automatically appear on your lab launcher page to start a notebook with that kernel.
User-defined kernels#
If the available modules in the pure module environment do not provide all Python packages that you need, then you can locally install these extra packages, followed by installing the corresponding Jupyter kernel either from a Python Conda environment, or from a Python virtual environment. For R, you may create your customized environment using Conda environments for R.
Note
User kernels are stored by default in ${VSC_HOME}/.local/share/jupyter/kernels.
To override this and store your kernel specifications in a non-default location,
you may drop the following line in your ${VSC_HOME}/.bashrc:
export XDG_DATA_HOME=${VSC_DATA}/.local/share
When the ${XDG_DATA_HOME} variable is set, subsequent kernel installations
(for both Python and R) will reside in ${XDG_DATA_HOME}/jupyter/kernels.
To remove a kernel, find and delete the corresponding folder inside the kernels
subdirectory.
We strongly advice against modifying the contents of this folder, unless you
are aware of the consequences.
Conda environments for Python#
Assuming you have created a Conda environment for Python, the corresponding kernel needs to be installed for use with JupyterLab.
First activate the Conda environment:
module load Mamba
source $EBROOTMAMBA/etc/profile.d/conda.sh
conda activate <env_name>
Next, install the ipykernel package and the kernel itself:
conda install ipykernel
python -m ipykernel install --user --env PYTHONPATH "" --name <env_name> --display-name <kernel_name>
These commands should be executed from a shell (e.g. using ‘Login (Server) Shell Access’), and only need to be done once for a given environment. When launching a new JupyterLab session, this kernel should then show up in the overview of available kernels, and as a new tile under the ‘Notebook’ section when opening a new launcher.
Notes
The minimum supported Python version is Python 3.7.
The minimum supported ipykernel version is ipykernel 6.19.2.
Tip
In case you encounter issues such as freezing or crashing JupyterLab sessions with a previously existing kernel, then reinstalling that kernel may help.
Virtual environments for Python#
You can use Python virtual environments to generate custom kernels for your notebooks. Virtual environments provide a layer of isolation allowing users to install additional Python packages on top of the software modules without conflicts.
Before using a virtual environment from the lab interface, consider the following requirements:
The chosen Python module needs to be in the list of ‘Toolchain and Python versions’ of the JupyterLab form (e.g.
2023a and Python/3.11.3-GCCcore-12.3.0).When using a virtual environment, the same software modules must be loaded as those that were loaded when creating it.
A virtual environment is only guaranteed to work in the same VSCdocCPU micro-architecture as the one it was created on.
Open the Terminal from your lab interface, ensuring that the requirements listed above are met.
Follow the instructions in Python virtual environments to create a new virtual environment and install any Python packages in it. Keep in mind that loading the Python module is not necessary as that is already done by the JupyterLab session. This new virtual environment can be placed anywhere you like in the storage of the cluster.
Example sequence of commands to create a new virtual environment in the directoryvenv-zen4#$ python3 -m venv venv-zen4 --system-site-packages $ source venv-zen4/bin/activate (venv-zen4) $ python3 -m pip install --upgrade pip (venv-zen4) $ python3 -m pip install <insert_cool_package>
Add your new virtual environment as a new Jupyter kernel (from the same terminal shell)
$ python3 -m ipykernel install --user --name=venv-zen4
A new launcher will appear in the lab interface to start notebooks using this new virtual environment

Launchers for default Python kernel and custom Python kernel from virtual environment#
To verify your setup, you can execute import sys; sys.executable in the new
kernel notebook, and the resulting path should point to the location of your
virtual environment.
Conda environments for R#
For R, you need both the jupyter_client and the irkernel Conda packages installed.
With the following command you can create the kernel:
Rscript -e 'IRkernel::installspec(name="<env_name>", displayname="<kernel_name>")'
Once the kernel is created, you will see it in the ‘Launcher’ menu. You can now start working in your own customized environment.
For more general information, please refer to the official JupyterLab documentation.
JupyterLab extensions#
JupyterLab extensions enhance or customize your JupyterLab session. You can find the list of available extensions in the extension tab on the left panel (puzzle piece icon) and you can enable or disable any of them.
Note
The JupyterLab extensions store is disabled because the available extensions in the store are not reviewed and may contain malicious or malfunctioning software. If you need any JupyterLab extension that is not yet available, please contact the site admins.
Software modules Lab extension#
The ‘Software Modules’ JupyterLab extension allows you to load additional software modules within JupyterLab without relaunching your JupyterLab session. This provides a more flexible alternative to loading modules via the ‘Pre-run Scriptlet’ in the resources form.
The ‘Software Modules’ Lab extension is enabled by default in your JupyterLab session. You can load software modules from the tab with a hexagon icon on the left panel of JupyterLab. This tab shows a long list of modules: the loaded modules at the top, and below that, a list of available modules.
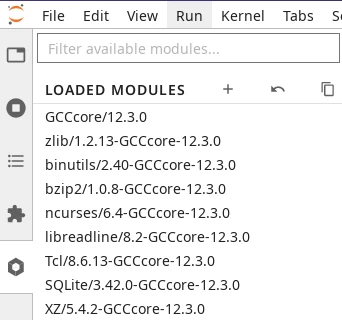
Software Modules Lab extension#
Upon launch, the list of loaded modules will already show some modules that have been loaded by JupyterLab itself. For example, you will always see a Python module loaded, which determines the Python version of the kernel used by your Python notebooks on this session.
Warning
Modules already loaded when your JupyterLab environment starts are necessary for the correct function of the lab and notebooks. They should not be unloaded.
Below the loaded modules, you will find the list of available modules that can be loaded on-demand. Move your mouse pointer to the right of the module name and a Load button will appear (see screenshot on the right).
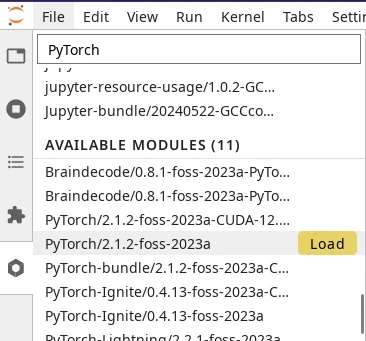
Loading a module in the Software Modules Lab extension#
Note
Any change to the list of loaded modules requires rebooting the kernel of
your open notebooks. After loading/unloading modules, click the kernel at the
top-right of the notebook toolbar, (default = Python 3 (ipykernel)) in the
screenshot below, and re-select your notebook kernel from the menu.
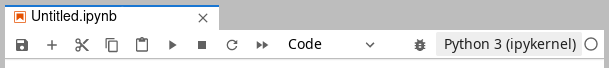
Notebook toolbar with default Python kernel#
Matplotlib Lab extension#
The matplotlib JupyterLab extension
(ipympl) enables the interactive features of matplotlib in your JupyterLab
session.
To enable the Lab extension, use the %matplotlib ipympl or %matplotlib
widget magic command. To ensure your plot is always shown, make sure to
generate a figure object before plotting, e.g. with plt.figure() or
plt.subplots().
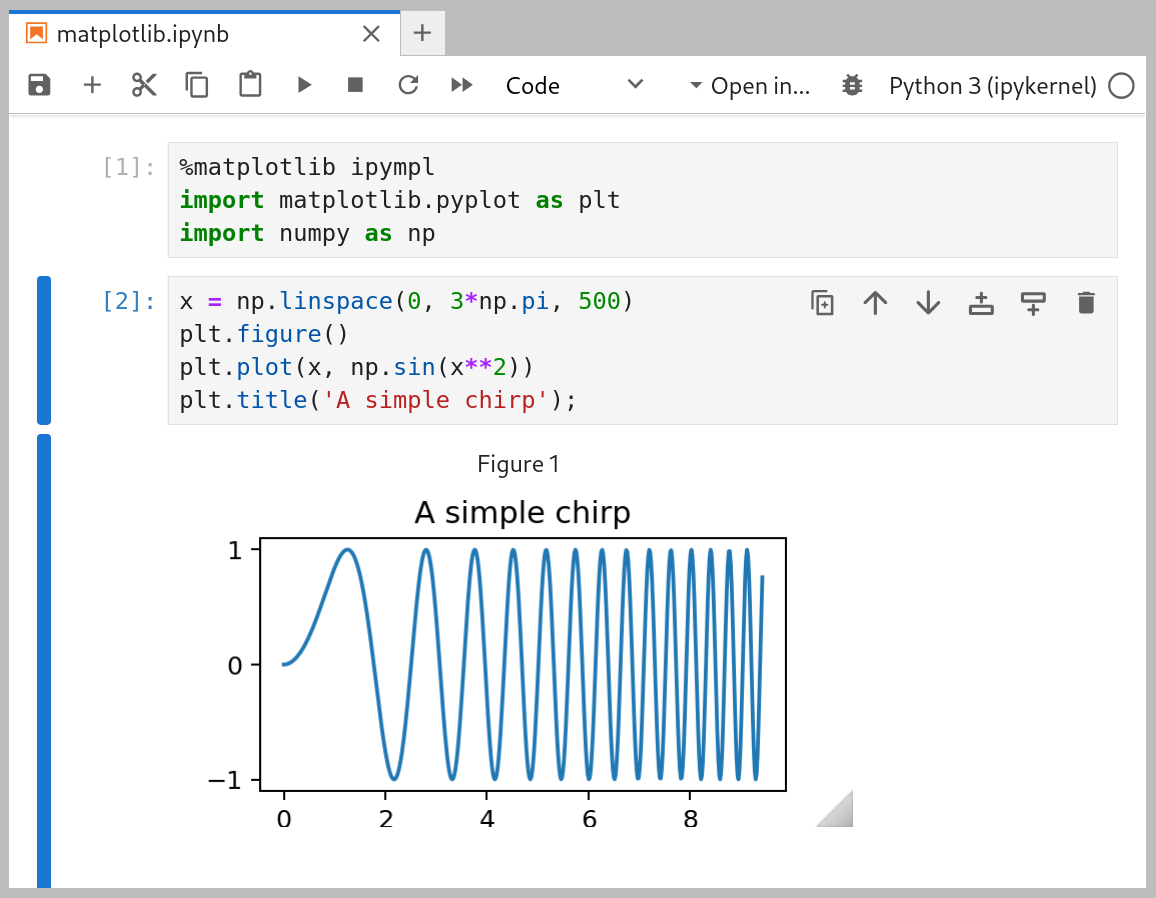
matplotlib Lab extension example#
Dask Lab extension#
The Dask JupyterLab extension (dask-labextension) provides tools to manage Dask clusters and embed Dask’s dashboard plots directly into your JupyterLab session.
In the resources form, tick the ‘Load the dask module’ checkbox to make sure the Dask Lab extension is loaded before starting JupyterLab.
In your new JupyterLab session, open a Jupyter notebook or start a new one.
In the ‘Settings’ menu, select ‘Auto-Start Dask’.
Click the Dask icon on the left side of the JupyterLab window.
In the ‘Clusters’ pane of the ‘Dask’ tab, click the ‘+New’ button to fire up a Dask cluster and connect your notebook to it. The default cluster is a LocalCluster, which is suited for single-node calculations.
You can now click any of the yellow-colored bars to open the corresponding Dask dashboard. Once opened, you can drag-and-drop it anywhere you want, and you can open multiple dashboards in the same session. Some useful Dask dashboards include ‘Cpu’, ‘Cluster Memory’, ‘Task Stream’, and ‘Workers’.
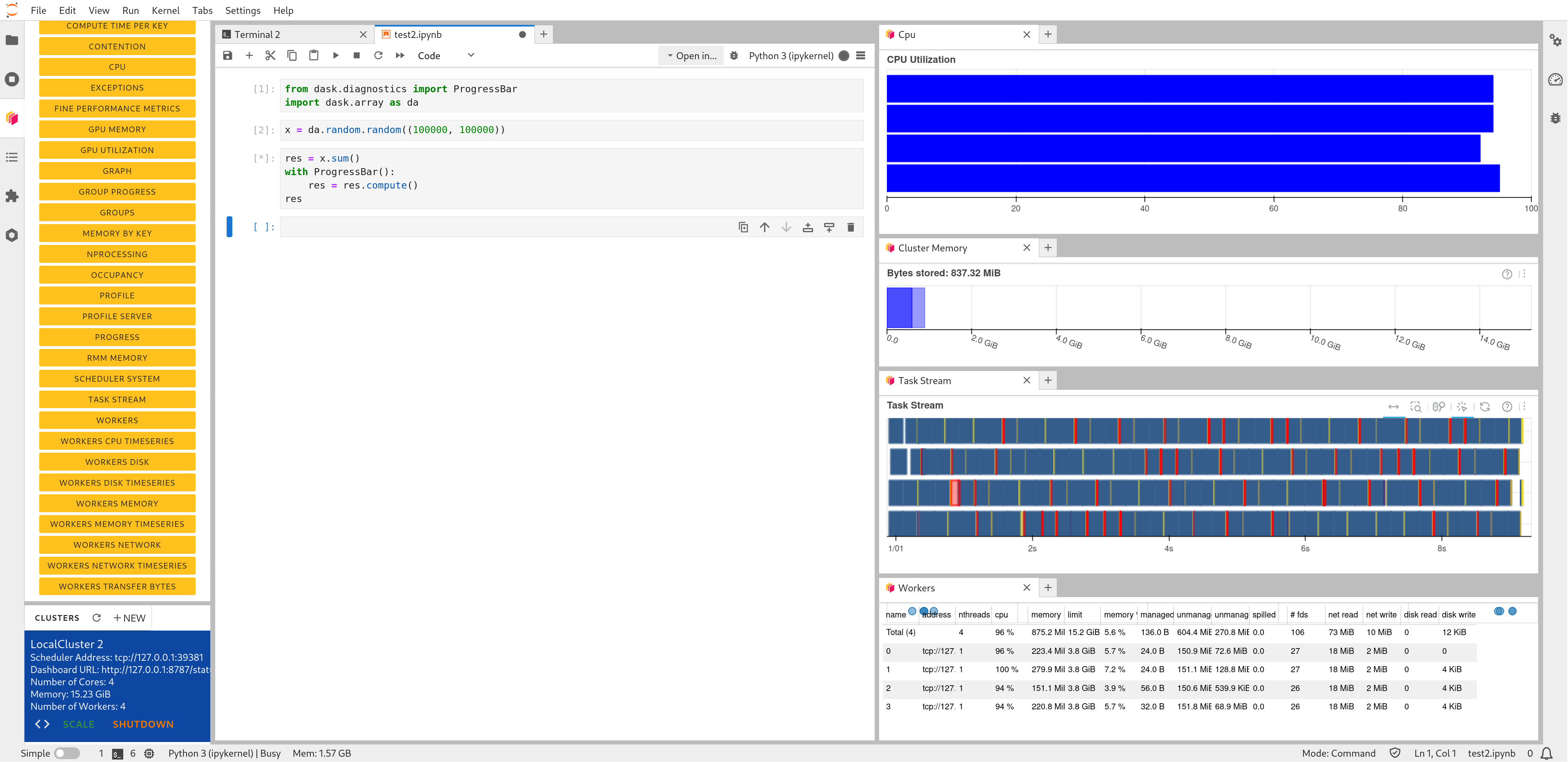
Dask Lab extension in action#
